
Metabolomics Laboratory Services & Fees
At NYU Langone’s Metabolomics Laboratory, research scientists and bioinformaticians use advanced technologies to perform sophisticated metabolome analyses. We customize our services according to the needs of your research project, whether global (untargeted) or targeted, at competitive rates.
Targeted projects are useful for testing a specific hypothesis, often with a large n-value. Global projects are useful for pilot studies or discovery-mode science. The two project types complement each other, with global projects leading to validation by targeted methods.
For targeted projects, we quantify the amount of one or more known or predetermined metabolites. For global projects, we detect whatever metabolites are present within a sample or look for differentially regulated metabolites between experimental groups.
Metabolomics Laboratory Fees
We offer custom services at competitive rates. Pricing depends on the specifications of your project and your institutional affiliation and is subject to change.
NYU Langone faculty and staff can use their Kerberos ID and password to log into iLab for more information on pricing. If you’re new to the system, we have provided iLab login instructions for NYU Langone Health, NYU, NYU College of Dentistry, and external users.
Example of a Global Metabolomics Project
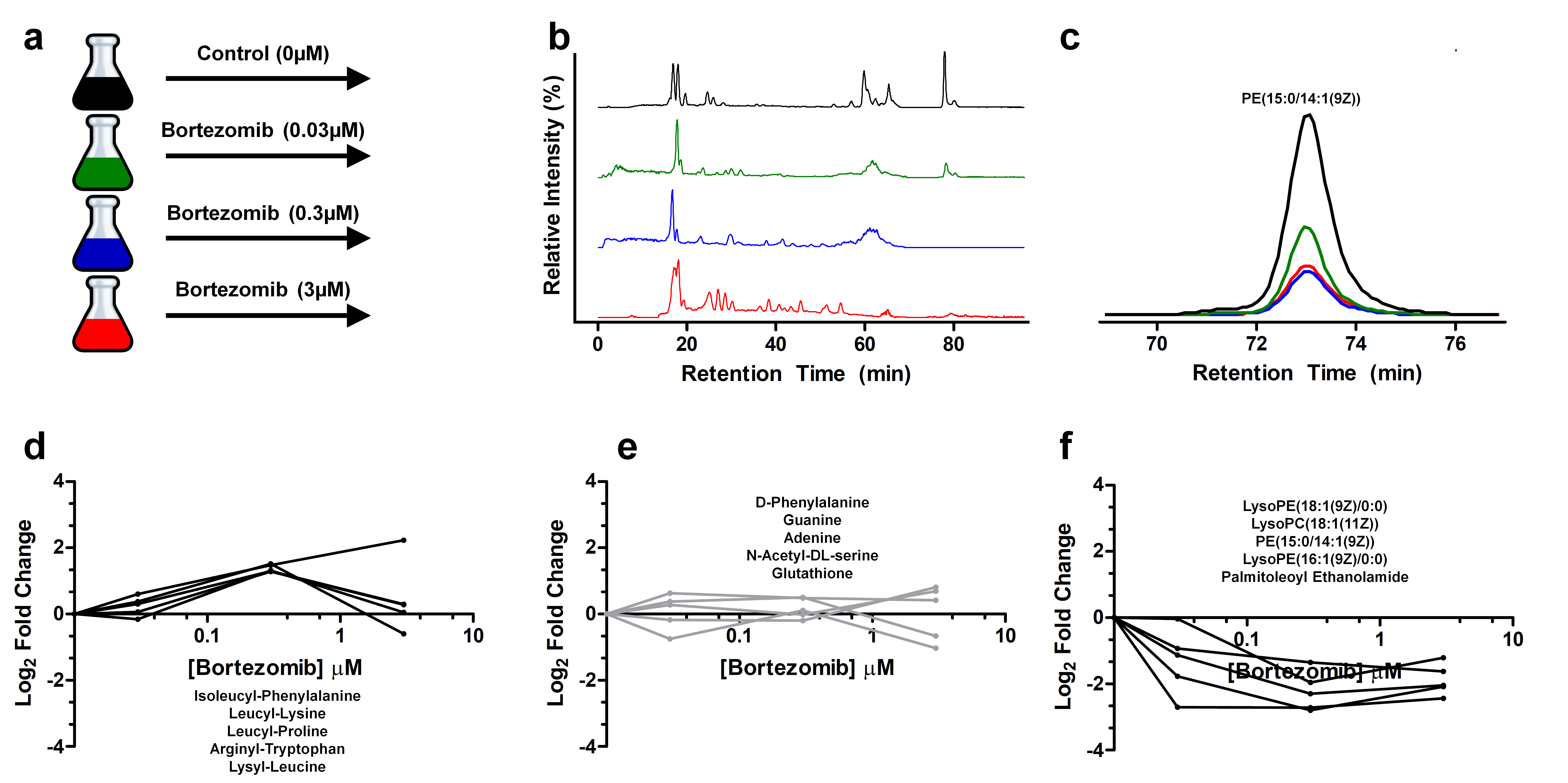
In this example project, we sought to determine the effect of the proteasome inhibitor bortezomib on the metabolic state of Saccharomyces cerevisiae. The experiment included a control group, which received no drug treatment, and several experimental groups, which received low, medium, and high treatment doses (panel a).
After treatment, we subjected the cells to a global metabolomics analysis using liquid chromatography–mass spectrometry, focusing on hydrophobic metabolites (panel b). We then used various bioinformatics tools to search for differentially regulated features. (For more information on these tools, see Gowda and colleagues in Analytical Chemistry; Wishart and colleagues in Nucleic Acids Research; and Jones and colleagues in bioRxiv.)
Overall, we detected approximately 6,000 peaks, 200 metabolite formulas, and 100 metabolite structures. One of these structures, a lipid metabolite in panel c, is shown to be downregulated with higher doses of bortezomib. Panels d, e, and f show a few of the upregulated, unchanged, and downregulated metabolites in the dataset, respectively.
From this pilot analysis, we independently recapitulated the drug mechanism by detecting incomplete proteasome degradation products as the most upregulated features. We also reported the novel finding that various phospholipids are dramatically downregulated after proteasome inhibition. These identified metabolites may serve as biomarkers of drug activity that can be verified with targeted experiments and potentially used to probe drug activity in large-scale experiments at a much lower cost.
We are developing new tools and resources to map this type of global data onto known metabolic pathways and to interface these results with genomics, RNA sequencing, proteomics, and other large datasets.
Compatible Sample Types
We have tested our global metabolomics pipeline in a wide range of sample types, from yeast cultures to clinical samples. If you have an unusual sample type, we can conduct pilot experiments to see if your system is compatible with our workflow.
Single-Cell Cultures
Compatible single-cell cultures include the following:
- yeast
- bacteria
- media supernatant
Mammalian Cell and Flow Cytometry–Purified Cultures
Compatible mammalian cell and flow cytometry–purified cultures include the following:
- HEK293
- myeloma
- T cells and T helper 17 cells
- primary neurons
- MDA-MB-231
- PA-TU-8988T
Tissue and Organ Samples
Compatible tissues and organ cells include the following:
- liver
- brain
- muscle
- heart
- kidney
- spleen
- lung
- bone marrow
Biofluids
Compatible biofluids include the following:
- blood
- plasma
- serum
- cerebrospinal fluid
- urine
- bile
- bronchoalveolar lavage
- saliva
- fecal
Whole Organisms
Compatible whole organisms include the following:
- Arabidopsis
- Drosophila
- Caenorhabditis elegans
Consulting and Grant Support
Our Metabolomics Laboratory can help you design your experiment and provide statistical analysis, such as power analysis, prior to sample collection. We also assist with grant applications and can provide letters of support, preliminary data if available, description of facilities, authentication of methods, biosketches, and more.
Please contact our director, Drew Jones, PhD, at drew.jones@nyumc.org to discuss your project before submitting samples.