
Transcriptional Regulation of Stem Cells
Investigators in the Dynlacht Lab use genome-wide methods including ChIP-seq and ChIP-on-chip to decipher the regulatory networks that control myogenic differentiation. Myogenesis is orchestrated through a series of transcriptional controls governed by myogenic regulatory factors (MRFs). MyoD, a basic helix-loop-helix (bHLH) transcription factor that binds sequence elements termed E-boxes, is the founding member of the MRF family which includes the closely related Myf5, myogenin, and MRF4 proteins. MRFs are key in the activation of the expression of genes that specify muscle.
The first steps in the myogenic transcriptional regulatory cascade involve expression of MyoD and Myf5, which subsequently leads to expression of myogenin and MEF2, promoting conversion of myoblasts to myotubes. MyoD collaborates with myogenin to regulate the expression of genes necessary for terminal differentiation. Myogenic differentiation proceeds through irreversible cell cycle arrest of precursor cells (myoblasts), followed by a gradual increase in expression of muscle function genes, leading to fusion of myoblasts into multinucleate myofibers in the animal. This process can be recapitulated in vitro, wherein myoblasts can be converted to myotubes with high efficiency in well-established models. In adult skeletal muscle, a pool of self-renewing stem cells (satellite cells) proliferate and differentiate in response to specific stimuli, such as injury or exercise.
Given the similarities between regeneration and the muscle differentiation program and the observation that deficiency of certain muscle regulatory factors (MRFs) leads to defects in muscle regeneration, we sought to identify the critical downstream targets of two MRFs, MyoD and the related MRF, myogenin, which is known to play a critical role in the later stages of muscle development. We used a combination of genome-wide approaches to identify physiological targets of MyoD1.
In addition, by combining chromatin immunoprecipitation and high throughput sequencing (ChIP-seq) and computational strategies in myoblasts and terminally differentiated myotubes, we identified the compendium of activating and repressive histone modifications that are deposited during myogenic differentiation.
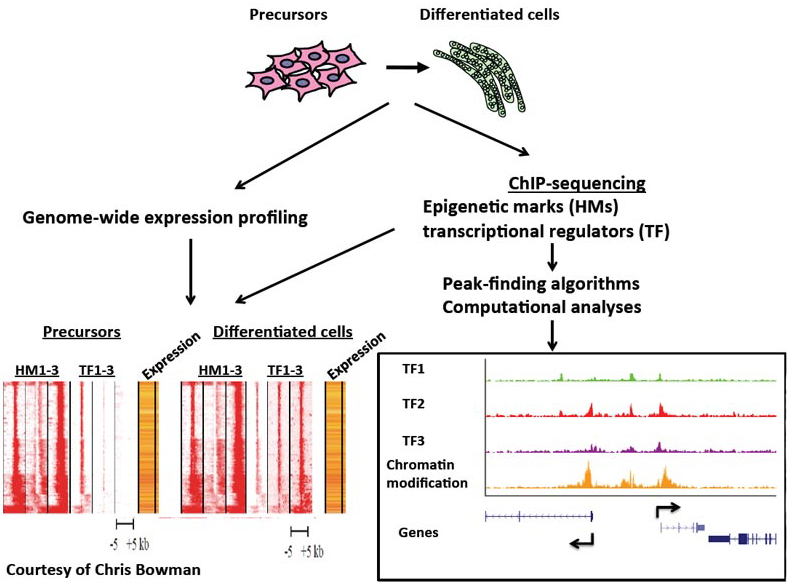
We have further identified the compendium of promoters and enhancers and found that MyoD1 is recruited to distal regulatory elements, where it helps to assemble transcriptional enhancers.
More recently, we have examined the recruitment of sequence-specific transcription factors, elongation factors, histone-modifying enzymes, and chromatin readers. By examining more than 30 histone modifications and transcriptional regulatory proteins, we have assembled one of the largest genome-wide datasets extant for a single tissue. These data have allowed us to begin dissecting a comprehensive network of key factors governing skeletal muscle differentiation.
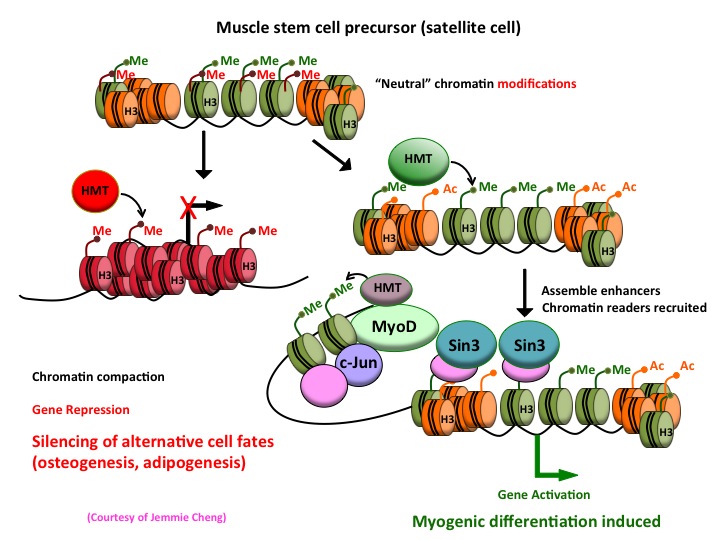
By investigating muscle stem cells, we hope to understand the epigenetic mechanisms that act as regulatory switches to determine mesodermal fates by silencing genes in certain lineages (fat and bone) while activating those in muscle. We hope that these studies will shed important new light on the myogenic transcriptional network and reveal novel insights into mesoderm development, muscle function, and regeneration, as well as disease.