HR is promoted by the action of recombinases, proteins that promote the formation of DNA joints that link homologous DNA molecules together. The eukaryotic recombinases are called Rad51 and Dmc1. Rad51 acts in somatic and meiotic cells while Dmc1 is active only in meiotic cells. One level of regulation of the HR process is through Rad51 activity. DNA helicases can affect the stability of the Rad51 presynaptic filament, a helical filament of Rad51 formed on ssDNA in preparation for strand invasion and DNA joint formation. Once a DNA joint is formed, Rad51 becomes associated with dsDNA. Rad51 must be removed to allow DNA polymerases to complete the recombination reactions. The Swi/Snf translocases Rad54 and Rdh54 can remove Rad51 from dsDNA and thus promote HR.
The Srs2 DNA helicase
The Srs2 protein is a 3’ - 5’ DNA helicase that has multiple functions in HR and repair. In HR, Srs2 has several distinct roles. It disrupts the Rad51 presynaptic filament and thus acts as an anti-recombinase, to ensure that DNA lesions are repaired in a nonrecombinational fashion. Interestingly, Srs2 also has pro-recombination roles: it promotes certain types of recombination events and after an induced double strand break, srs2 mutants show low viability and never seem to recover from the DNA damage checkpoint-imposed cell cycle arrest. The anti-recombination role of Srs2 stems from its ability to disrupt the Rad51 presynaptic filament, thereby preventing inappropriate recombination events from occurring.
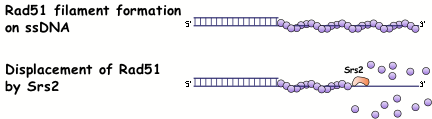
Our current studies on Srs2 involve understanding how Srs2 contributes to HR through its interactions with Rad51 and other recombination factors. We are examining srs2 mutants that are defective in binding to Rad51 to see what functions are reduced or defective and what functions remain normal. These studies will reveal the different domains of Srs2 and the complexes in which Srs2 can act.
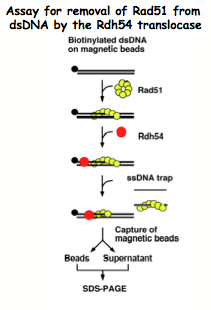
Swi/Snf translocases Rad54 and Rdh54
Both Rad54 and Rdh54 function in HR at several steps. We are interested in the steps that involve an interaction with Rad51. We are delineating the Rad51 interaction domains and determining the role these play in HR. Other areas of study are focused on identifying additional proteins that interact with Rad54 and Rdh54 and determining new roles for Rad54 and Rdh54 in chromosome structure and segregation. We are also studying new Swi/Snf family members for a role in HR and Rad51 interaction.
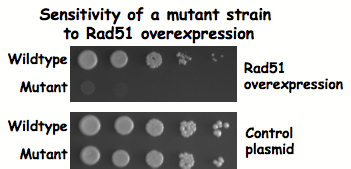
Rad51 overexpression
Since Rad51 is only transiently associated with DNA during HR, this suggests that there must be a balance between free Rad51 protein and the factors that remove it from ssDNA and dsDNA. We are exploring how this balance is maintained by overexpressing Rad51 and disrupting the balance. This situation also mimics what occurs in many kinds of cancer cells, where Rad51 is increased in expression with resulting drug and radiation resistance and increased genomic instability. We have identified mutants that are exquisitely sensitive to Rad51 overexpression, a condition called synthetic dosage lethality, and are doing screens against the entire yeast gene deletion collection to find new sensitive mutants.
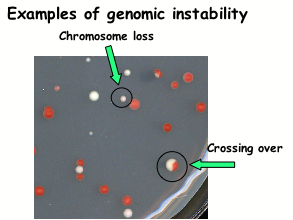
Genomic stability assays
We are using diploid cells with genetically marked chromosomes to study the different types of events that can lead to genomic instability; chromosome loss, chromosome rearrangements, chromosome missegregation, recombination, mutation and other types of eprevent genomic instability from spontaneous DNA damage.